1. Christ, Patrick Ferdinand, et al. "Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2016.
Goal : Segmentation of liver and its lesions in CT abdomen images
Dataset : 3DIRCADb dataset (https://www.ircad.fr/research/3d-ircadb-01/)
- 3D CT scans for 20 people with hepatic tumors, where 15 volumes have hepatic tumors in the liver.
Preprocessing : in slice-wise
1) Hounsfield unit values windowed to range [-100, 400] to exclude irrelevant organs and organs.
2) Increase contrast through histogram equalization
3) Augment data by applying translation, rotation and addition of gaussian noise
-> Increased training dataset of 22,693 image slices.
Model : Cascaded FCNs(CFCNs) based on UNet architecture and dense 3D conditional random fields(CFRs)
1) Train the first FCN to segment the liver as ROI input to second FCN.
2) Train the second FCN to segment lesions from the predicted liver ROIs of 1).
3) Refine the result using a dense 3D CRF

Performance : Dice scores over 94%, computation times below 100s per volume.
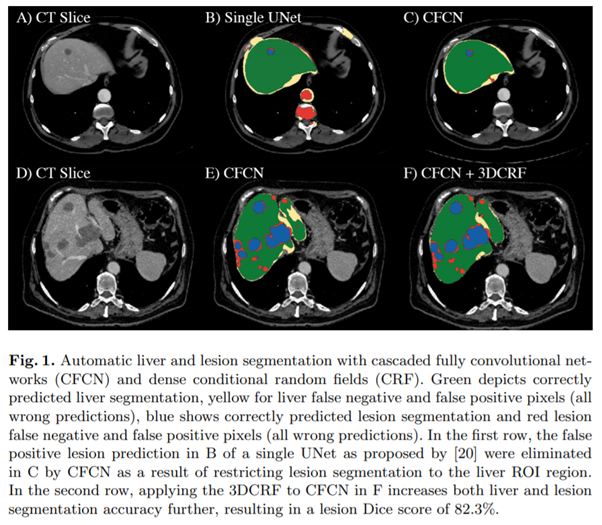
- Green : correctly predicted liver segmentation
- Yellow : liver FN and FP (wrong predictions)
- Blue : correctly predicted lesion segmentation
- Red : lesion FN and FP (wrong predictions)
- B(single UNet (2015))에서의 FP lesion 부분이 C에서 사라짐.
- E에서 3DCRF를 적용한 F에서 accuracy가 증가함.(both liver & lesion)

2. Li, Xiaomeng, et al. "H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes." IEEE transactions on medical imaging 37.12 (2018): 2663-2674.
Problem : 2D FCN은 3D의 공간적인 정보를 전부 담아낼 수 없고, 3D FCN은 high computational cost.
Solution : Hybrid Densely connected UNet (H-DenseUNet), which consists of a 2-D DenseUNet for efficiently extracting intra-slice features and a 3-D counterpart for hierarchically aggregating volumetric context under the spirit of the auto-context algorithm for liver and tumor segmentation.
Dataset : MICCAI 2017 Liver Tumor Segmentation Challenge(LiTS) and 3DIRCADb dataset
- LiTS : 131 and 70 contrast-enhanced 3D abdominal CT scans
Preprocessing :
1) HU to [-200, 250] to remove the irrelevant details
2) For coarse liver segmentation, resample images with the same resolution 0.69 * 0.69 * 1.0 mm3
3) For lesion segmentation, use image with original resolution
Model Architecture : employed cascaded learning strategy
1) simple ResNet architecture to get a quick but coarse segmentation of liver as ROI
2) Using ROI from 1), H-DenseUNet probes intra-slice and inter-slice features through a 2D DenseUNet and 3D counterpart.
3) Hybrid Feature Fusion (HFF) layer for accurate segmentation


Performance :



3. https://www.kakaobrain.com/blog/48 (2018)
- MICCAI가 주관한 3D 의료영상 분할 알고리즘 개발 대회인 ‘medical decathlon’에서 2등 수상
Goal : 서로 다른 10종의 의료영상에 대해 일정 수준 이상의 정확도와 속도로 이미지를 segmentation하는 범용 모델

* 의료영상에서의 범용 모델의 필요성
1) 전체 데이터셋 개수의 부족
2) 클래스 불균형
3) 다중 레이블의 존재
4) Multimodality image 학습의 어려움
Model : 2단계 3D UNet
- 첫 번째 UNet은 전체 이미지 중 중요한 부분을 crop함.
- 두 번째 UNet은 첫 번째 UNet이 crop한 부분을 input으로 받아 segmentation함.
